class: center, middle, inverse, title-slide # Reproducible science: Module 3 ## Dealing with data: Tidyverse ### <a href="dossa@xtbg.org.cn">Gbadamassi G.O. Dossa</a> ### Xishuangbanna Tropical Botanical Garden, XTBG-CAS ### 2021/09/25 (updated: 2022-06-27) --- class: center # Acknowledgements The content of this module are based on materials from: .pull-right[  ] .pull-right[ [olivier gimenez's materials](https://oliviergimenez.github.io/) ] --- class: center # What is tidyverse and advantages? .left[ "A framework for managing data that aims at making the cleaning and preparing steps [muuuuuuuch] easier" (Julien Barnier). Main characteristics of a tidy dataset: - the dataset is [tibble](https://tibble.tidyverse.org/); - measured varaiable as a column; - an observation represents a row with each value is in a different cell. [**tidyverse**](https://www.tidyverse.org/) consits of a compilation of r packages for data analysis. ]  <img src="img/tidyverse.png" width="10%" style="display: block; margin: auto 0 auto auto;" /> --- class: center # Recognizing a tidy dataset  .left[Is this a tidy data?] -- .left[No] --- class: center # Recognizing a tidy dataset  .left[Is this a tidy data?] -- .left[No] --- class: center # Recognizing a tidy dataset  .left[Is this a tidy data?] -- .left[No] --- class: center # Recognizing a tidy dataset  .left[Is this a tidy data?] -- .left[Yes] --- class: center # Tidyverse: Multiple r packages well compiled .left[Allows using a consistent format for which powerful tools work. Makes data manipulation pretty natural - [ggplot2](https://ggplot2-book.org/) - visualizing stuff; - [dplyr](https://dplyr.tidyverse.org/), [tidyr](https://tidyr.tidyverse.org/) - data manipulation; - [purrr](https://purrr.tidyverse.org/) - advanced programming; - [readr](https://readr.tidyverse.org/) - import data; - [tibble](https://tibble.tidyverse.org/) - improved data.frame format; - [forcats](https://forcats.tidyverse.org/) - working with factors; - [stringr](https://stringr.tidyverse.org/) - working with chain of characters. ] --- class: center # Simplified flowchart of data science? .left[Any data analysis follows this typical flow: 1. Import data; 2. Clean data; 3. Exploratory analysis. A cycle between: - Visualization; - modeling; - Transformation 4. Communicate] <small>If these steps happen at multiple software then errors are highly inevitable.</small> <div class="figure" style="text-align: center"> <img src="img/workflow in data analysis.png" alt="Reproducibilit equals effecient use of time" width="60%" /> <p class="caption">Reproducibilit equals effecient use of time</p> </div> --- class: center # Tidyverse saves: same flowchart in tidyverse <div class="figure" style="text-align: center"> <img src="img/workflow data analysis within tidyverse.png" alt="Reproducibilit equals effecient use of time" width="80%" /> <p class="caption">Reproducibilit equals effecient use of time</p> </div> --- class: center # Practice in tidyverse “Use twitter to predict citation rate”  .left[ We will use an existing data supporting the [above publication](https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0166570) to learn some functions within **tidyverse**. ] --- class: center # Import data .left[ readr::read_csv function: - creates tibbles instead of data.frame; - no names to rows; - allows column names with special characters (see next slide); - more clever on screen display than w/ data.frames (see next slide); - no partial matching on column names; - warning if attempt to access unexisting column; - is incredibly fast. ] --- class: center # Import data .left[ ```r # Set the url from where to download the data url<-"https://doi.org/10.1371/journal.pone.0166570.s001" # name the file to be downloaded and save as destfile object destfile <- "twitter_cit_data.csv" # Apply download.file function in R to download from url download.file(url, destfile) library(tidyverse) ``` ``` ## Warning: package 'ggplot2' was built under R version 4.1.1 ``` ``` ## Warning: package 'readr' was built under R version 4.1.1 ``` ```r # Read the data file with read_csv() and save with name "citations_raw" *citations_raw<-read_csv(file="twitter_cit_data.csv") head(citations_raw) ``` ] --- class: center # Import data .left[ ```r citations_raw ``` ``` ## # A tibble: 1,599 x 12 ## `Journal identity` `5-year journal im~ `Year published` Volume Issue Authors ## <chr> <dbl> <dbl> <dbl> <chr> <chr> ## 1 Ecology Letters 16.7 2014 17 12 Morin e~ ## 2 Ecology Letters 16.7 2014 17 12 Jucker ~ ## 3 Ecology Letters 16.7 2014 17 12 Calcagn~ ## 4 Ecology Letters 16.7 2014 17 11 Segre e~ ## 5 Ecology Letters 16.7 2014 17 11 Kaufman~ ## 6 Ecology Letters 16.7 2014 17 10 Nasto e~ ## 7 Ecology Letters 16.7 2014 17 10 Tschirr~ ## 8 Ecology Letters 16.7 2014 17 9 Barnech~ ## 9 Ecology Letters 16.7 2014 17 9 Pinto-S~ ## 10 Ecology Letters 16.7 2014 17 9 Clough ~ ## # ... with 1,589 more rows, and 6 more variables: Collection date <chr>, ## # Publication date <chr>, Number of tweets <dbl>, Number of users <dbl>, ## # Twitter reach <dbl>, Number of Web of Science citations <dbl> ``` ] --- class: center # Tidy/transform: Rename columns .left[ To rename columns, use function *rename()* new_name=old_name ```r citations_temp <- rename(citations_raw, journal = 'Journal identity', impactfactor = '5-year journal impact factor', pubyear = 'Year published', colldate = 'Collection date', pubdate = 'Publication date', nbtweets = 'Number of tweets', woscitations = 'Number of Web of Science citations') head(citations_temp,5,6) ``` ``` ## # A tibble: 5 x 12 ## journal impactfactor pubyear Volume Issue Authors colldate pubdate nbtweets ## <chr> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl> ## 1 Ecology ~ 16.7 2014 17 12 Morin e~ 2/1/2016 9/16/2~ 18 ## 2 Ecology ~ 16.7 2014 17 12 Jucker ~ 2/1/2016 10/13/~ 15 ## 3 Ecology ~ 16.7 2014 17 12 Calcagn~ 2/1/2016 10/21/~ 5 ## 4 Ecology ~ 16.7 2014 17 11 Segre e~ 2/1/2016 8/28/2~ 9 ## 5 Ecology ~ 16.7 2014 17 11 Kaufman~ 2/1/2016 8/28/2~ 3 ## # ... with 3 more variables: Number of users <dbl>, Twitter reach <dbl>, ## # woscitations <dbl> ``` ] --- class: center # Tidy: Clean up column names .left[ To clean columns, use function *clean_names()* from the package janitor from it will fill space in column names by "_". ```r janitor::clean_names(citations_raw) ``` ``` ## # A tibble: 1,599 x 12 ## journal_identity x5_year_journal_impa~ year_published volume issue authors ## <chr> <dbl> <dbl> <dbl> <chr> <chr> ## 1 Ecology Letters 16.7 2014 17 12 Morin et ~ ## 2 Ecology Letters 16.7 2014 17 12 Jucker et~ ## 3 Ecology Letters 16.7 2014 17 12 Calcagno ~ ## 4 Ecology Letters 16.7 2014 17 11 Segre et ~ ## 5 Ecology Letters 16.7 2014 17 11 Kaufman e~ ## 6 Ecology Letters 16.7 2014 17 10 Nasto et ~ ## 7 Ecology Letters 16.7 2014 17 10 Tschirren~ ## 8 Ecology Letters 16.7 2014 17 9 Barnechi ~ ## 9 Ecology Letters 16.7 2014 17 9 Pinto-San~ ## 10 Ecology Letters 16.7 2014 17 9 Clough et~ ## # ... with 1,589 more rows, and 6 more variables: collection_date <chr>, ## # publication_date <chr>, number_of_tweets <dbl>, number_of_users <dbl>, ## # twitter_reach <dbl>, number_of_web_of_science_citations <dbl> ``` ] --- class: center # Tidy: Create and modify columns .left[ The well known function to create and modify columns is *mutate()*, This function takes first the tibble names, the new_name= what you want to do to old column. ```r citations <- mutate(citations_temp, journal = as.factor(journal)) #Pay attention that I store in "citations" citations ``` ``` ## # A tibble: 1,599 x 12 *## journal impactfactor pubyear Volume Issue Authors colldate pubdate nbtweets *## <fct> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl> ## 1 Ecology~ 16.7 2014 17 12 Morin e~ 2/1/2016 9/16/2~ 18 ## 2 Ecology~ 16.7 2014 17 12 Jucker ~ 2/1/2016 10/13/~ 15 ## 3 Ecology~ 16.7 2014 17 12 Calcagn~ 2/1/2016 10/21/~ 5 ## 4 Ecology~ 16.7 2014 17 11 Segre e~ 2/1/2016 8/28/2~ 9 ## 5 Ecology~ 16.7 2014 17 11 Kaufman~ 2/1/2016 8/28/2~ 3 ## 6 Ecology~ 16.7 2014 17 10 Nasto e~ 2/2/2016 7/28/2~ 27 ## 7 Ecology~ 16.7 2014 17 10 Tschirr~ 2/2/2016 8/6/20~ 6 ## 8 Ecology~ 16.7 2014 17 9 Barnech~ 2/2/2016 6/17/2~ 19 ## 9 Ecology~ 16.7 2014 17 9 Pinto-S~ 2/2/2016 6/12/2~ 26 ## 10 Ecology~ 16.7 2014 17 9 Clough ~ 2/2/2016 7/17/2~ 44 ## # ... with 1,589 more rows, and 3 more variables: Number of users <dbl>, ## # Twitter reach <dbl>, woscitations <dbl> ``` ] --- # Tidy: Create and modify columns .left[ Check now the levels of journal variable ```r levels(citations$journal) ``` ``` ## [1] "Animal Conservation" "Conservation Letters" ## [3] "Diversity and Distributions" "Ecological Applications" ## [5] "Ecology" "Ecology Letters" ## [7] "Evolution" "Evolutionary Applications" ## [9] "Fish and Fisheries" "Functional Ecology" ## [11] "Global Change Biology" "Global Ecology and Biogeography" ## [13] "Journal of Animal Ecology" "Journal of Applied Ecology" ## [15] "Journal of Biogeography" "Limnology and Oceanography" ## [17] "Mammal Review" "Methods in Ecology and Evolution" ## [19] "Molecular Ecology Resources" "New Phytologist" ``` ] --- class: center # Piping: Make your manipulations easier Piping was borrowed from other languages, got incorporated into R after a question in [Pipe question](https://stackoverflow.com/questions/8896820/how-to-implement-fs-forward-pipe-operator-in-r) in 2012. Pipe which is the bar "|" on your keyboard. --- class: center # Omelette: Base r approach You need to do complicated programming: create multiple intermediate objects; embed, needs some understanding of coding and is prone to errors. --- class: center # Omelette: Piping approach .left[Simpler programming using piping. Piping consists of: taking results from previous function as a starting point of a new function;less prone to errors and consume less memory. ] --- class: center # Example of piping .left[ Take the tibble "citations_raw" **then** rename some columns then the new tibble containing the renamed tibble and *then* convert the column "journal" from current class ("character") to factor. ```r citations_raw %>% * rename(journal = 'Journal identity', impactfactor = '5-year journal impact factor', pubyear = 'Year published', colldate = 'Collection date', pubdate = 'Publication date', nbtweets = 'Number of tweets', woscitations = 'Number of Web of Science citations') %>% * mutate(journal = as.factor(journal)) ``` ] .left[ Please notice every time I say **"then"** this is equal to **"%>%"**. ] --- class: center # Naming final object of pipe .left[ ```r *citations <- citations_raw %>% rename(journal = 'Journal identity', impactfactor = '5-year journal impact factor', pubyear = 'Year published', colldate = 'Collection date', pubdate = 'Publication date', nbtweets = 'Number of tweets', woscitations = 'Number of Web of Science citations') %>% mutate(journal = as.factor(journal)) head(citations) ``` ``` ## # A tibble: 6 x 12 ## journal impactfactor pubyear Volume Issue Authors colldate pubdate nbtweets ## <fct> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl> ## 1 Ecology ~ 16.7 2014 17 12 Morin e~ 2/1/2016 9/16/2~ 18 ## 2 Ecology ~ 16.7 2014 17 12 Jucker ~ 2/1/2016 10/13/~ 15 ## 3 Ecology ~ 16.7 2014 17 12 Calcagn~ 2/1/2016 10/21/~ 5 ## 4 Ecology ~ 16.7 2014 17 11 Segre e~ 2/1/2016 8/28/2~ 9 ## 5 Ecology ~ 16.7 2014 17 11 Kaufman~ 2/1/2016 8/28/2~ 3 ## 6 Ecology ~ 16.7 2014 17 10 Nasto e~ 2/2/2016 7/28/2~ 27 ## # ... with 3 more variables: Number of users <dbl>, Twitter reach <dbl>, ## # woscitations <dbl> ``` ] --- class: center # Naming final object of pipe 2 .left[ ```r citations_raw %>% rename(journal = 'Journal identity', impactfactor = '5-year journal impact factor', pubyear = 'Year published', colldate = 'Collection date', pubdate = 'Publication date', nbtweets = 'Number of tweets', woscitations = 'Number of Web of Science citations') %>% * mutate(journal = as.factor(journal))-> citations2 head(citations2) ``` ``` ## # A tibble: 6 x 12 ## journal impactfactor pubyear Volume Issue Authors colldate pubdate nbtweets ## <fct> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl> ## 1 Ecology ~ 16.7 2014 17 12 Morin e~ 2/1/2016 9/16/2~ 18 ## 2 Ecology ~ 16.7 2014 17 12 Jucker ~ 2/1/2016 10/13/~ 15 ## 3 Ecology ~ 16.7 2014 17 12 Calcagn~ 2/1/2016 10/21/~ 5 ## 4 Ecology ~ 16.7 2014 17 11 Segre e~ 2/1/2016 8/28/2~ 9 ## 5 Ecology ~ 16.7 2014 17 11 Kaufman~ 2/1/2016 8/28/2~ 3 ## 6 Ecology ~ 16.7 2014 17 10 Nasto e~ 2/2/2016 7/28/2~ 27 ## # ... with 3 more variables: Number of users <dbl>, Twitter reach <dbl>, ## # woscitations <dbl> ``` ] --- class: center # Pipe synthax .left[ - *Verb(Subject,Complement)* replaced by *Subject %>% Verb(Complement)*; - No need to name unimportant intermediate variables; - Clear syntax (readability). ] .left[ If you want you can first write what you want to accomplished in a text with "then" as step wise, then code it by replace "then" by the pipe with its operator "%>%" of course. <img src="img/pipinglogo.png" width="10%" style="display: block; margin: auto 0 auto auto;" /> ] --- class: center, middle, inverse # Other functions in Tidyverse --- class: center # Select columns .left[*select()* is the function one uses to select different variables i a tibble. You just need to remember that it follows a pipe operator (%>%), and it takes the name of columns one desires to select. ```r citations %>% select(journal, impactfactor, nbtweets) ``` ``` ## # A tibble: 1,599 x 3 ## journal impactfactor nbtweets ## <fct> <dbl> <dbl> ## 1 Ecology Letters 16.7 18 ## 2 Ecology Letters 16.7 15 ## 3 Ecology Letters 16.7 5 ## 4 Ecology Letters 16.7 9 ## 5 Ecology Letters 16.7 3 ## 6 Ecology Letters 16.7 27 ## 7 Ecology Letters 16.7 6 ## 8 Ecology Letters 16.7 19 ## 9 Ecology Letters 16.7 26 ## 10 Ecology Letters 16.7 44 ## # ... with 1,589 more rows ``` ] --- class: center # Drop columns or deselect variables .left[ The opposite of selecting, which is deselecting. One just need to be more logical in the writing. Would you like to guess? ] -- .left[ ```r citations %>% * select(-Volume, -Issue, -Authors) ``` ``` ## # A tibble: 1,599 x 9 ## journal impactfactor pubyear colldate pubdate nbtweets `Number of user~ ## <fct> <dbl> <dbl> <chr> <chr> <dbl> <dbl> ## 1 Ecology Le~ 16.7 2014 2/1/2016 9/16/2014 18 16 ## 2 Ecology Le~ 16.7 2014 2/1/2016 10/13/20~ 15 12 ## 3 Ecology Le~ 16.7 2014 2/1/2016 10/21/20~ 5 4 ## 4 Ecology Le~ 16.7 2014 2/1/2016 8/28/2014 9 8 ## 5 Ecology Le~ 16.7 2014 2/1/2016 8/28/2014 3 3 ## 6 Ecology Le~ 16.7 2014 2/2/2016 7/28/2014 27 23 ## 7 Ecology Le~ 16.7 2014 2/2/2016 8/6/2014 6 6 ## 8 Ecology Le~ 16.7 2014 2/2/2016 6/17/2014 19 18 ## 9 Ecology Le~ 16.7 2014 2/2/2016 6/12/2014 26 23 ## 10 Ecology Le~ 16.7 2014 2/2/2016 7/17/2014 44 42 ## # ... with 1,589 more rows, and 2 more variables: Twitter reach <dbl>, ## # woscitations <dbl> ``` ] --- class: center # Split a column in several columns .left[separate is the function used to split a column into several of course you need to indicate what symbol is the separator (e.g., space, -, /, etc.). ```r head(citations$pubdate) ``` ``` ## [1] "9/16/2014" "10/13/2014" "10/21/2014" "8/28/2014" "8/28/2014" ## [6] "7/28/2014" ``` ```r citations %>% select(journal, impactfactor, nbtweets, pubdate)%>% separate(pubdate,c('month','day','year'),'/') ``` ``` ## # A tibble: 1,599 x 6 ## journal impactfactor nbtweets month day year ## <fct> <dbl> <dbl> <chr> <chr> <chr> ## 1 Ecology Letters 16.7 18 9 16 2014 ## 2 Ecology Letters 16.7 15 10 13 2014 ## 3 Ecology Letters 16.7 5 10 21 2014 ## 4 Ecology Letters 16.7 9 8 28 2014 ## 5 Ecology Letters 16.7 3 8 28 2014 ## 6 Ecology Letters 16.7 27 7 28 2014 ## 7 Ecology Letters 16.7 6 8 6 2014 ## 8 Ecology Letters 16.7 19 6 17 2014 ## 9 Ecology Letters 16.7 26 6 12 2014 ## 10 Ecology Letters 16.7 44 7 17 2014 ## # ... with 1,589 more rows ``` ] --- class: center # Transform column in date format .left[Many of us work with ecological data that record date, and we find it hard to keep these on readable format in R. Within, tidyverse there is a package that specially deals with date formatting variables/columns. The package is called [**lubridate**](https://lubridate.tidyverse.org/). ```r library(lubridate) citations %>% mutate(pubdate = mdy(pubdate), colldate = mdy(colldate))%>% select(journal,impactfactor, nbtweets, pubdate, colldate) ``` ``` ## # A tibble: 1,599 x 5 ## journal impactfactor nbtweets pubdate colldate ## <fct> <dbl> <dbl> <date> <date> ## 1 Ecology Letters 16.7 18 2014-09-16 2016-02-01 ## 2 Ecology Letters 16.7 15 2014-10-13 2016-02-01 ## 3 Ecology Letters 16.7 5 2014-10-21 2016-02-01 ## 4 Ecology Letters 16.7 9 2014-08-28 2016-02-01 ## 5 Ecology Letters 16.7 3 2014-08-28 2016-02-01 ## 6 Ecology Letters 16.7 27 2014-07-28 2016-02-02 ## 7 Ecology Letters 16.7 6 2014-08-06 2016-02-02 ## 8 Ecology Letters 16.7 19 2014-06-17 2016-02-02 ## 9 Ecology Letters 16.7 26 2014-06-12 2016-02-02 ## 10 Ecology Letters 16.7 44 2014-07-17 2016-02-02 ## # ... with 1,589 more rows ``` ] --- class: center # For easy date format manipulation .left[Check out ?lubridate::lubridate for more functions ```r library(lubridate) citations %>% mutate(pubdate = mdy(pubdate), pubyear2 = year(pubdate))%>% select(journal,impactfactor, pubdate, colldate, pubyear2) ``` ``` ## # A tibble: 1,599 x 5 ## journal impactfactor pubdate colldate pubyear2 ## <fct> <dbl> <date> <chr> <dbl> ## 1 Ecology Letters 16.7 2014-09-16 2/1/2016 2014 ## 2 Ecology Letters 16.7 2014-10-13 2/1/2016 2014 ## 3 Ecology Letters 16.7 2014-10-21 2/1/2016 2014 ## 4 Ecology Letters 16.7 2014-08-28 2/1/2016 2014 ## 5 Ecology Letters 16.7 2014-08-28 2/1/2016 2014 ## 6 Ecology Letters 16.7 2014-07-28 2/2/2016 2014 ## 7 Ecology Letters 16.7 2014-08-06 2/2/2016 2014 ## 8 Ecology Letters 16.7 2014-06-17 2/2/2016 2014 ## 9 Ecology Letters 16.7 2014-06-12 2/2/2016 2014 ## 10 Ecology Letters 16.7 2014-07-17 2/2/2016 2014 ## # ... with 1,589 more rows ``` ] --- class: center, middle, inverse # Join tables together --- class: center # Join two tables .left[ Joining tables are the correspondents of merge function in base R. There is a great tutorial to all sort of joining in tidyverse made available by [Garrick Aden-Buie](https://www.garrickadenbuie.com/project/tidyexplain/). The joining of tables can be categorized into several types. However, we will only study the following: - Inner join; - Left join; - Right join; - Semi join; - Union join; - Anti join. ] --- class: center # Inner join  --- class: center # Left join  --- class: center # Right join  --- class: center # Semi join  --- class: center # Union join 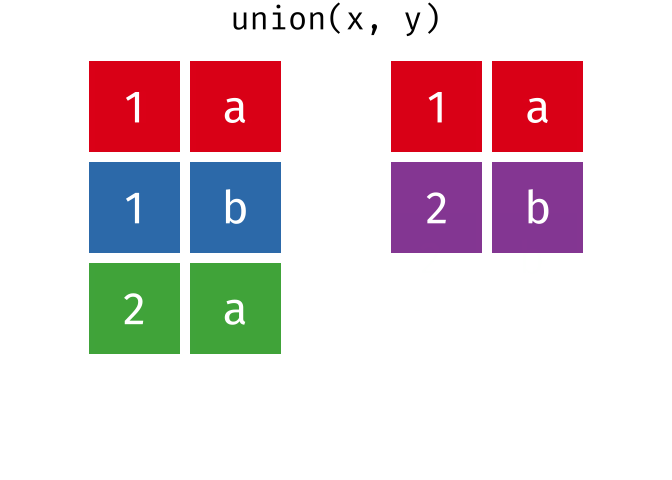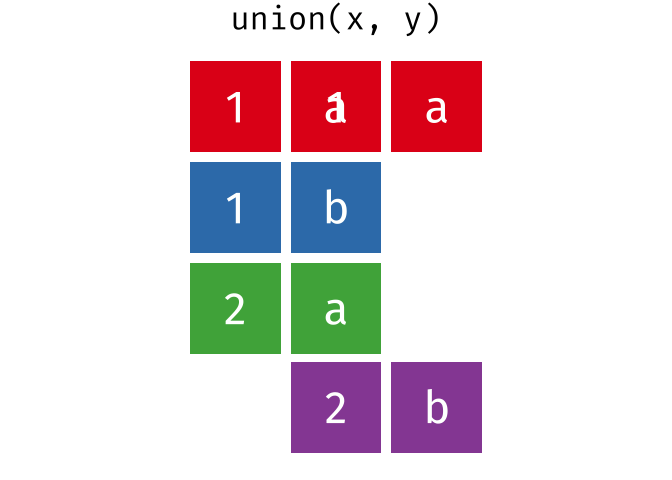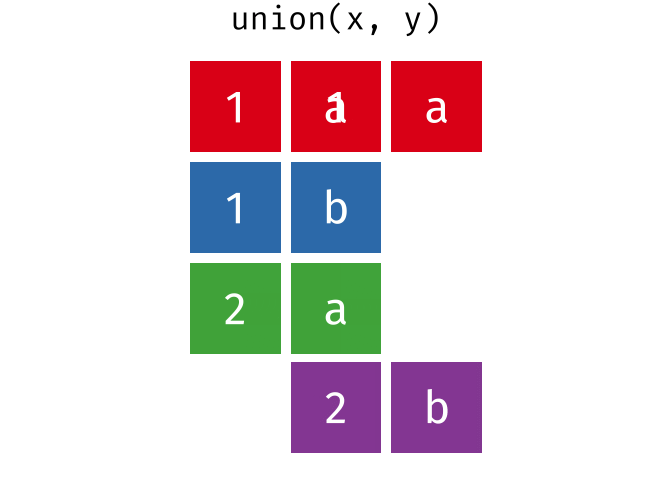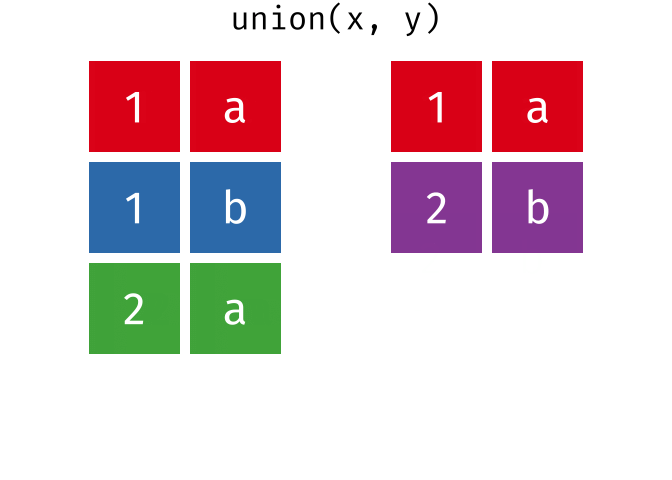 --- class: center # Anti join 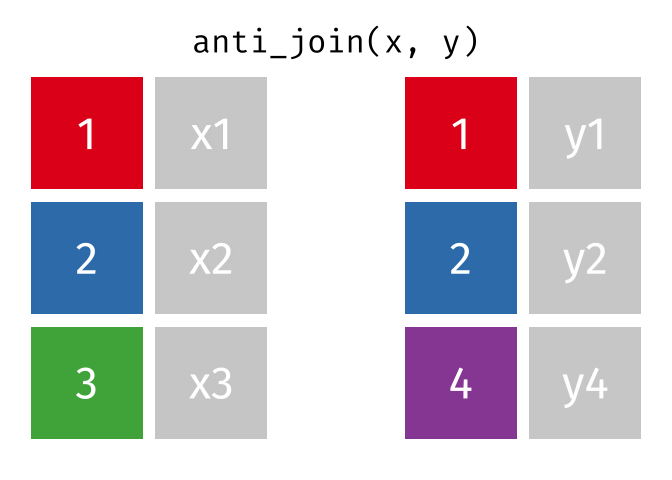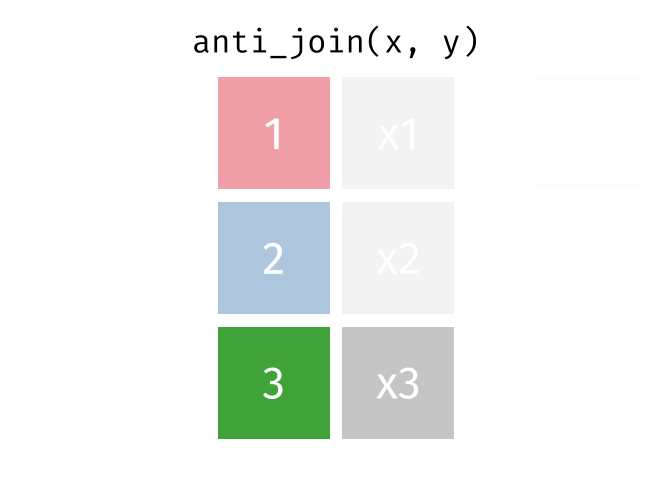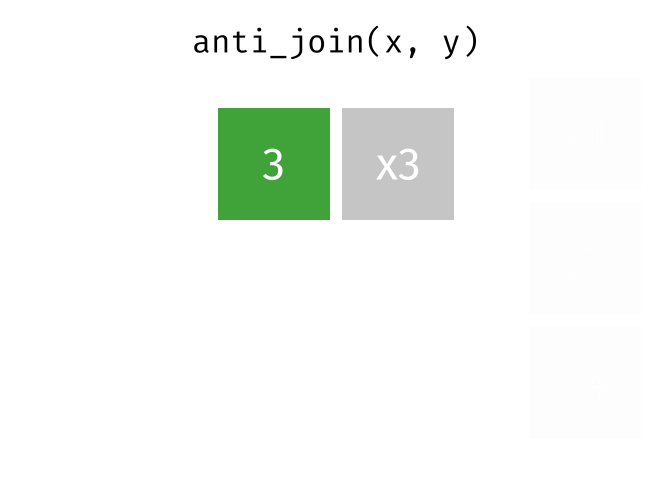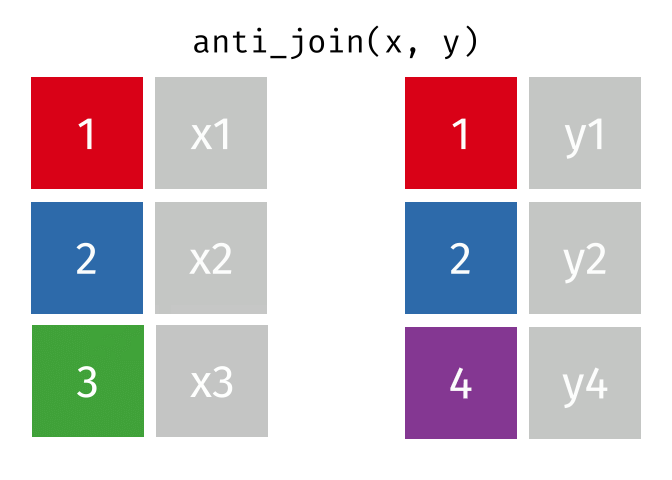 --- class: center, middle, inverse # Character manipulation --- class: center # Select rows of papers with > 3 authors .left[ ```r citations %>% *#str_detect() detect characters in a given column filter(str_detect(Authors,'et al')) ``` ``` ## # A tibble: 1,280 x 12 ## journal impactfactor pubyear Volume Issue Authors colldate pubdate nbtweets ## <fct> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl> ## 1 Ecology~ 16.7 2014 17 12 Morin e~ 2/1/2016 9/16/2~ 18 ## 2 Ecology~ 16.7 2014 17 12 Jucker ~ 2/1/2016 10/13/~ 15 ## 3 Ecology~ 16.7 2014 17 12 Calcagn~ 2/1/2016 10/21/~ 5 ## 4 Ecology~ 16.7 2014 17 11 Segre e~ 2/1/2016 8/28/2~ 9 ## 5 Ecology~ 16.7 2014 17 11 Kaufman~ 2/1/2016 8/28/2~ 3 ## 6 Ecology~ 16.7 2014 17 10 Nasto e~ 2/2/2016 7/28/2~ 27 ## 7 Ecology~ 16.7 2014 17 10 Tschirr~ 2/2/2016 8/6/20~ 6 ## 8 Ecology~ 16.7 2014 17 9 Barnech~ 2/2/2016 6/17/2~ 19 ## 9 Ecology~ 16.7 2014 17 9 Pinto-S~ 2/2/2016 6/12/2~ 26 ## 10 Ecology~ 16.7 2014 17 9 Clough ~ 2/2/2016 7/17/2~ 44 ## # ... with 1,270 more rows, and 3 more variables: Number of users <dbl>, ## # Twitter reach <dbl>, woscitations <dbl> ``` ] --- class: center # Select columns with rows of papers with > 3 authors .left[ ```r citations %>% filter(str_detect(Authors,'et al')) %>% select(Authors) ``` ``` ## # A tibble: 1,280 x 1 ## Authors ## <chr> ## 1 Morin et al ## 2 Jucker et al ## 3 Calcagno et al ## 4 Segre et al ## 5 Kaufman et al ## 6 Nasto et al ## 7 Tschirren et al ## 8 Barnechi et al ## 9 Pinto-Sanchez et al ## 10 Clough et al ## # ... with 1,270 more rows ``` ] --- class: center # Select columns with rows of papers with < 3 authors .left[ ```r citations %>% filter(!str_detect(Authors,'et al')) %>% ##! for saying "not". select(Authors) ``` ``` ## # A tibble: 319 x 1 ## Authors ## <chr> ## 1 Neutle and Thorne ## 2 Kellner and Asner ## 3 Griffin and Willi ## 4 Gremer and Venable ## 5 Cavieres ## 6 Haegman and Loreau ## 7 Kearney ## 8 Locey and White ## 9 Quintero and Weins ## 10 Lesser and Jackson ## # ... with 309 more rows ``` ] --- class: center # Select authors of columns with rows of papers with < 3 authors .left[ ```r citations %>% filter(!str_detect(Authors,'et al')) %>% ##! for saying "not". pull(Authors) %>% head(10) ``` ``` ## [1] "Neutle and Thorne" "Kellner and Asner" "Griffin and Willi" ## [4] "Gremer and Venable" "Cavieres" "Haegman and Loreau" ## [7] "Kearney" "Locey and White" "Quintero and Weins" ## [10] "Lesser and Jackson" ``` ] --- class: center # Rows of papers with less than 3 authors in journal with IF < 5 .left[ ```r citations %>% filter(!str_detect(Authors,'et al'), impactfactor < 5) ``` ``` ## # A tibble: 77 x 12 ## journal impactfactor pubyear Volume Issue Authors colldate pubdate nbtweets ## <fct> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl> ## 1 Molecul~ 4.9 2014 14 6 Gautier 2/27/20~ 5/14/2~ 2 ## 2 Molecul~ 4.9 2014 14 5 Gambel~ 2/27/20~ 3/7/20~ 7 ## 3 Molecul~ 4.9 2014 14 4 Kekkon~ 2/27/20~ 3/10/2~ 4 ## 4 Molecul~ 4.9 2014 14 3 Bhatta~ 2/27/20~ 12/8/2~ 0 ## 5 Molecul~ 4.9 2014 14 1 Christ~ 2/28/20~ 10/25/~ 0 ## 6 Molecul~ 4.9 2013 13 4 Villar~ 2/28/20~ 5/2/20~ 0 ## 7 Molecul~ 4.9 2013 13 4 Wang 2/28/20~ 4/25/2~ 0 ## 8 Molecul~ 4.9 2012 12 1 Joly 2/28/20~ 9/7/20~ 3 ## 9 Animal ~ 3.21 2014 17 6 Plavsic 2/9/2016 4/17/2~ 9 ## 10 Animal ~ 3.21 2014 17 Suppl~ Knox a~ 2/11/20~ 11/13/~ 1 ## # ... with 67 more rows, and 3 more variables: Number of users <dbl>, ## # Twitter reach <dbl>, woscitations <dbl> ``` ] --- class: center # Convert words to lowercase .left[ ```r citations %>% * mutate(authors_lowercase = str_to_lower(Authors)) %>% select(authors_lowercase) ``` ``` ## # A tibble: 1,599 x 1 ## authors_lowercase ## <chr> ## 1 morin et al ## 2 jucker et al ## 3 calcagno et al ## 4 segre et al ## 5 kaufman et al ## 6 nasto et al ## 7 tschirren et al ## 8 barnechi et al ## 9 pinto-sanchez et al ## 10 clough et al ## # ... with 1,589 more rows ``` ] --- class: center # Remove all spaces in variable names .left[ ```r citations%>% mutate(journal = str_remove_all(journal," ")) %>% select(journal) %>% unique() %>% head(5) ``` ``` ## # A tibble: 5 x 1 ## journal ## <chr> ## 1 EcologyLetters ## 2 GlobalChangeBiology ## 3 GlobalEcologyandBiogeography ## 4 MolecularEcologyResources ## 5 DiversityandDistributions ``` ] --- class: center, middle, inverse # Basic exploratory data analysis --- class: center # Count () This helps to count the number of occurrences. .left[ ```r citations %>% count(journal, sort = TRUE) ## Embedded sorting within count() ``` ``` ## # A tibble: 20 x 2 ## journal n ## <fct> <int> ## 1 New Phytologist 144 ## 2 Ecology 108 ## 3 Evolution 108 ## 4 Global Change Biology 108 ## 5 Global Ecology and Biogeography 108 ## 6 Journal of Biogeography 108 ## 7 Ecology Letters 106 ## 8 Diversity and Distributions 105 ## 9 Animal Conservation 102 ## 10 Methods in Ecology and Evolution 90 ## 11 Evolutionary Applications 74 ## 12 Functional Ecology 54 ## 13 Journal of Animal Ecology 54 ## 14 Journal of Applied Ecology 54 ## 15 Limnology and Oceanography 54 ## 16 Molecular Ecology Resources 54 ## 17 Conservation Letters 53 ## 18 Ecological Applications 48 ## 19 Fish and Fisheries 36 ## 20 Mammal Review 31 ``` ] --- class: center # Count() for multiple variables .left[ ```r citations %>% count(journal, pubyear) ``` ``` ## # A tibble: 59 x 3 ## journal pubyear n ## <fct> <dbl> <int> ## 1 Animal Conservation 2012 18 ## 2 Animal Conservation 2013 18 ## 3 Animal Conservation 2014 66 ## 4 Conservation Letters 2012 17 ## 5 Conservation Letters 2013 18 ## 6 Conservation Letters 2014 18 ## 7 Diversity and Distributions 2012 36 ## 8 Diversity and Distributions 2013 33 ## 9 Diversity and Distributions 2014 36 ## 10 Ecological Applications 2012 24 ## # ... with 49 more rows ``` ] --- class: center # Count sum of tweets per journal .left[ ```r citations %>% count(journal, wt = nbtweets, sort = TRUE) ``` ``` ## # A tibble: 20 x 2 ## journal n ## <fct> <dbl> ## 1 Ecology Letters 1538 ## 2 Animal Conservation 1268 ## 3 Journal of Applied Ecology 1012 ## 4 Methods in Ecology and Evolution 699 ## 5 Global Change Biology 613 ## 6 Conservation Letters 542 ## 7 New Phytologist 509 ## 8 Global Ecology and Biogeography 379 ## 9 Ecology 335 ## 10 Evolution 335 ## 11 Journal of Animal Ecology 323 ## 12 Fish and Fisheries 261 ## 13 Evolutionary Applications 238 ## 14 Journal of Biogeography 209 ## 15 Diversity and Distributions 200 ## 16 Mammal Review 166 ## 17 Functional Ecology 155 ## 18 Molecular Ecology Resources 139 ## 19 Ecological Applications 125 ## 20 Limnology and Oceanography 0 ``` ] --- class: center # Group variables to compute stats [summarise()] .left[ ```r citations %>% group_by(journal) %>% summarise(avg_tweets = mean(nbtweets)) ``` ``` ## # A tibble: 20 x 2 ## journal avg_tweets ## <fct> <dbl> ## 1 Animal Conservation 12.4 ## 2 Conservation Letters 10.2 ## 3 Diversity and Distributions 1.90 ## 4 Ecological Applications 2.60 ## 5 Ecology 3.10 ## 6 Ecology Letters 14.5 ## 7 Evolution 3.10 ## 8 Evolutionary Applications 3.22 ## 9 Fish and Fisheries 7.25 ## 10 Functional Ecology 2.87 ## 11 Global Change Biology 5.68 ## 12 Global Ecology and Biogeography 3.51 ## 13 Journal of Animal Ecology 5.98 ## 14 Journal of Applied Ecology 18.7 ## 15 Journal of Biogeography 1.94 ## 16 Limnology and Oceanography 0 ## 17 Mammal Review 5.35 ## 18 Methods in Ecology and Evolution 7.77 ## 19 Molecular Ecology Resources 2.57 ## 20 New Phytologist 3.53 ``` ] --- class: center # Order stuff [arrange()] .left[ ```r citations %>% group_by(journal) %>% summarise(avg_tweets = mean(nbtweets)) %>% * # decreasing order but (without desc for increasing) arrange(desc(avg_tweets))-> arrangedat head(arrangedat, 10) ``` ``` ## # A tibble: 10 x 2 ## journal avg_tweets ## <fct> <dbl> ## 1 Journal of Applied Ecology 18.7 ## 2 Ecology Letters 14.5 ## 3 Animal Conservation 12.4 ## 4 Conservation Letters 10.2 ## 5 Methods in Ecology and Evolution 7.77 ## 6 Fish and Fisheries 7.25 ## 7 Journal of Animal Ecology 5.98 ## 8 Global Change Biology 5.68 ## 9 Mammal Review 5.35 ## 10 New Phytologist 3.53 ``` ] --- class: center # Work on several columns [dplyr:::across()]  --- class: center # Compute mean across multiple variables .left[ ```r citations %>% group_by(journal) %>% * summarize(across(where(is.numeric), mean)) ``` ``` ## # A tibble: 20 x 8 ## journal impactfactor pubyear Volume nbtweets `Number of user~ `Twitter reach` ## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 Animal~ 3.21 2013. 16.5 12.4 9.71 28345. ## 2 Conser~ 6.4 2013. 6.02 10.2 8.85 23234. ## 3 Divers~ 5.4 2013 19 1.90 1.77 2350. ## 4 Ecolog~ 5.06 2013 23 2.60 2.5 5727. ## 5 Ecology 6.16 2013 94 3.10 2.87 6176. ## 6 Ecolog~ 16.7 2013. 16.0 14.5 14.0 44748. ## 7 Evolut~ 5.25 2013 67 3.10 2.93 7762. ## 8 Evolut~ 4.6 2013. 6.05 3.22 3.07 13185. ## 9 Fish a~ 8.1 2013 14 7.25 6.19 12097. ## 10 Functi~ 5.28 2013 27 2.87 2.74 3809. ## 11 Global~ 8.7 2013 19 5.68 4.94 9652. ## 12 Global~ 7.18 2013 22 3.51 3.15 8995. ## 13 Journa~ 5.32 2013. 81.9 5.98 5.59 36112. ## 14 Journa~ 5.93 2013 50 18.7 15.8 43839. ## 15 Journa~ 4.59 2013 40 1.94 1.86 11632. ## 16 Limnol~ 4.4 2013 58 0 0 0 ## 17 Mammal~ 4.3 2013. 43.0 5.35 4.39 10485. ## 18 Method~ 7.39 2013. 4.2 7.77 7.14 15867. ## 19 Molecu~ 4.9 2013 13 2.57 2.24 19844. ## 20 New Ph~ 7.8 2013 198. 3.53 3.15 6340. ## # ... with 1 more variable: woscitations <dbl> ``` ] --- class: center # Tidying tibbles [wide(), long()] 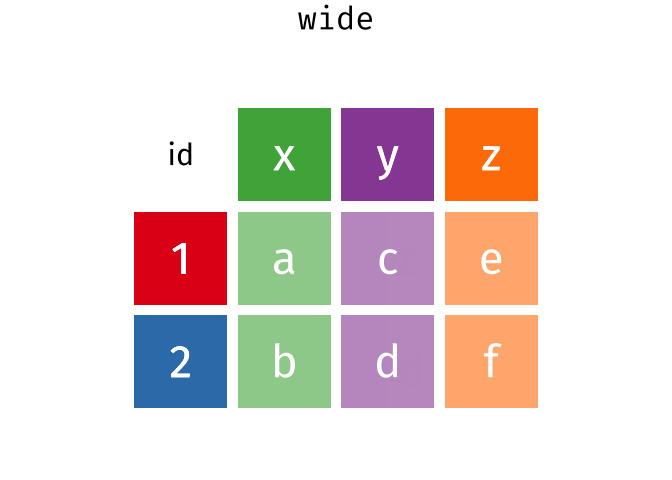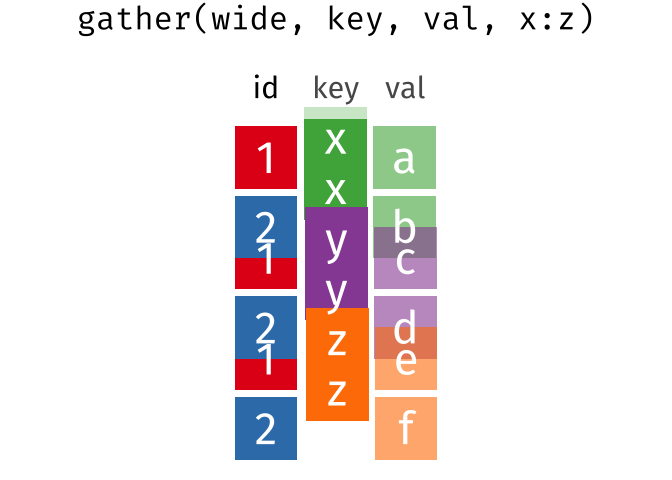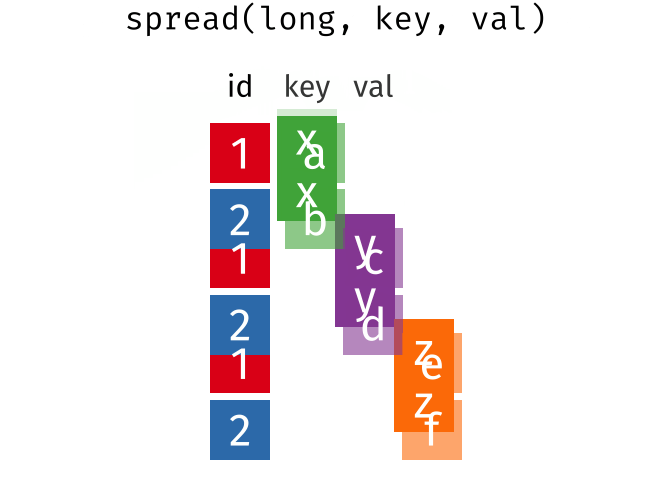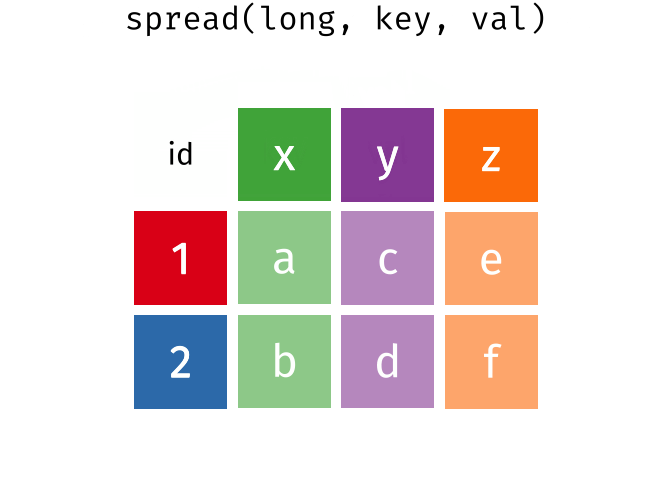 --- class: center # Data manipulation with tidyverse: in depth study .left[ Learn the tidyverse: books, workshops and online courses Selection of books: - [R for Data Science](https://www.tidyverse.org/learn/) and [Advanced R](http://adv-r.had.co.nz/); - [Tidy Tuesdays videos](https://www.youtube.com/user/safe4democracy/videos) by D. Robinson; - Material of the [stat545](https://stat545.com/) course on Data wrangling, exploration, and analysis with R at the University of British Columbia; - List of best R packages (with their description) on data import, [wrangling and visualization](https://www.computerworld.com/article/2921176/great-r-packages-for-data-import-wrangling-visualization.html). ] --- class: center, middle # Thank you for listening! Any questions now or email me at [**dossa@xtbg.org.cn**](http://people.ucas.edu.cn/~Dossa?language=en) Slides created via the R package [**xaringan**](https://github.com/yihui/xaringan). The chakra comes from [remark.js](https://remarkjs.com), [**knitr**](https://yihui.org/knitr/), and [R Markdown](https://rmarkdown.rstudio.com).